Presentation of my dissertation work
This week, I presented my dissertation work at the American Medical Informatics Association conference in Washington, DC. The entire panel (4 papers) on ontology mapping took place, aptly enough, in the Hilton's Map Room.
Ever wondered what happens when two papers on a panel have the same title? I inadvertently found out when Olivier Bodenreider and I independently gave the same title to our papers on mapping mouse anatomy to human anatomy: "Of Mice and Men", and both were accepted. So the first and third papers on the panel were each entitled "Of Mice and Men", and when Mark Musen introduced the panel, he quipped that we would retire afterward for refreshments to the Steinbeck room.
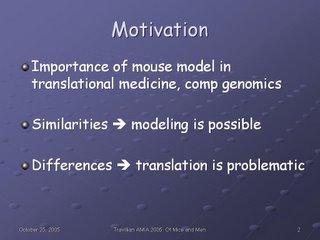
My talk presented my dissertation work, which addresses a very important problem: the accurate determination and representation of anatomical correspondences across species, in this case, human and mouse.

The importance of this problem has been well-documented; in fact, many of the people present in the audience had written on just that topic. Mice, as well as many other species, are used as model organisms for understanding disease in humans, and for deciphering the genome. They can serve this purpose because of the similarities between them and us, and it is these similarities which make comparative medicine possible. At the same time, the differences between us create issues in the interpretation and translation of results in these species to humans: treatments that cure cancer in mice don't necessarily do so in people, and there are reports of cardioprotective effects of Vioxx in mice, rats, and marmosets, which did not predict the harmful effects that drug would have in humans, just to name two examples. The effects of these similarities and differences emphasize the importance of sound and complete modeling of these structures across species, in order to support correct reasoning about the implications.
Because of the importance of the mouse as a model for human medicine and genomics, the Mouse Models of Human Cancer Consortium has identified and prioritized 11 types of cancer to consolidate murine information on. The cancers, listed on this slide, are each associated with specific anatomical sites, which is where our anatomy ontology and information system come in.

Our system is designed to answer user queries about the similarities and differences in human and mouse anatomy in selected sites taken from those identified from the MMHCC. The sites identified are the ovary, cervix, and mammary gland of the female mouse, the prostate of the male mouse, and the lung, shared by males and females.

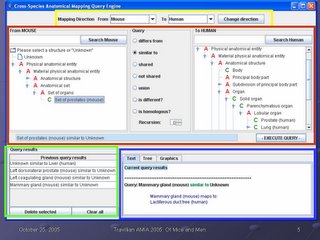
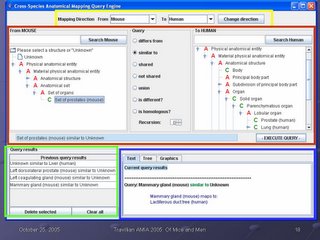
This is a screen shot of our application. There are four main areas, which are outlined on the screen.
The yellow box outlines the query direction. Users can choose to ask queries about the human compared to the mouse, or about the mouse compared to the human, as they prefer. In this way, the interface conforms to the user preference. However, as outline in my earlier MS thesis, the query is bidirectional: the user can ask the question in either direction and get the same result. In other words, the queries "how does the human prostate differ from the mouse prostate?" and "how does the mouse prostate differ from the human prostate?" return the same information. The user does not need to be concerned that the order of the query has any informational significance; it is only a user interface accommodation to the user's preference.
The green box at the bottom left is where previous queries and their outcomes are stored. A session can be cumulative in the sense that information can be gathered, and that information then used in later queries. In this way, our application builds on the work done earlier in our research group on Emily, a query interface to the Foundational Model of Anatomy. Emily also stores results of previous queries for reuse; as this was a most useful feature, we incorporated it into our system.
The blue box at the bottom left is where the current query results are returned. In the eaxmple in this slide, the user has just asked what structure in the human corresponds to the mammary gland in the mouse, and the system has returned the information "Mammary gland (mouse) maps to lactiferous duct tree (human)". What is displayed in this area is always the results from the last query submitted.
The red box in the center of the screen is where a new query is formed, and will make up the bulk of this talk. It consists of a "from" hierarchy (can be either mouse or human), a "to" hierarchy (either mouse or human), and a set of possible queries in the middle that can be selected by radio buttons. The text box below is populated as the user forms her query: in this way, she 1) does not have to remember our syntax to form a query, and 2) gets immediate feedback from the text box whether the query is what she wants, in time to change it or submit it. In the example in the slide, the user has selected "Set of prostates (mouse) similar to Unknown (human)" to ask "what structure(s) in the human correspond to the set of mouse prostates?", and the query is ready to submit by clicking on the Execute Query button.
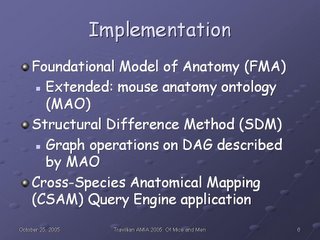
We implemented our system in the following way, described in more detail in my MS thesis and our AMIA 2003 paper: first, we used the Foundational Model of Anatomy as a template to build a partial ontology of selected mouse organs, an ontology which we shall refer to as the Mouse Anatomy Ontology (MAO).
We used the Structural Difference Method (outlined in my thesis) to perform graph operations on the directed acyclic graphs (DAGs) described by the FMA (human) and MAO (mouse).
Finally, we developed a user interface and query engine to present the result sets oobtained by the SDM to the user.
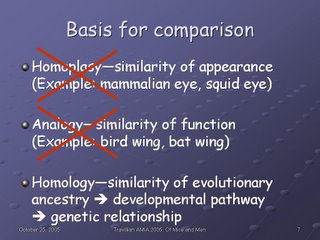
In order to determine the mappings, we had to establish what structure in one species was similar to what structure in the other species. Traditionally, when biologists and comparative anatomists have referred to "similarity", there are three different aspects that they take into account:
Homoplasy, or similarity of appearance: the mammalian eye and the squid eye appear similar to use--they "look like" eyes, which is the homoplasy. Yet structurally and developmentally, they are very different from each other. We do not model homoplasy in our model.
Analogy, or similarity of function: the bird wing and the bat wing both serve the purpose of flight, yet they evolved at different times, using different structures in the respective forelimbs involved. We do not model analogy in our model.
Homology, or similarity of evolutionary ancestry: while homologous structures may or may not exhibit homoplasy and/or analogy, it is their shared evolutionary ancestry--which we can get at via understanding of their developmental pathways, which in turn sheds light on the genetic relationships involved--that we concern ourselves with in our modeling of similarity and difference.
Because it is the homologies we are modeling, there is a natural tie-in to the Foundational Model of Anatomy ontology. As we see in the slide, by definition, Anatomical entity--a fundamental unit of the FMA--is a Material anatomical entity which has inherent 3D shape; is generated by coordinated expression of the organism's own structural genes; and its parts are spatially related to one another in patterns determined by coordinated gene expression.
The existence of the entity--coordinated by the structural genes--is what ties the FMA Anatomical Taxonomy (AT) component to homology. The spatial relationships among its parts--determined by coordinated gene expression--are what ties the FMA Anatomical Spatial Abstraction (ASA) component to homology.
A further component of the FMA, the Anatomical Transformation Abstraction (ATA), is also tied in to the developmental pathways aspect of homology; however, that is outside the scope of my dissertation, and will not be pursued here.

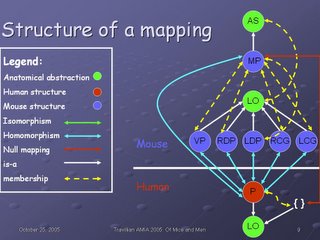
This slide shows the structure of a mapping, specifically the mapping between the human and mouse prostates. The red circle (P) represents the human prostate; the blue circles represent the mouse prostates ventral prostate (VP), right dorsolateral prostate (RDP), left dorsolateral prostate (LDP), right coagulating gland (RCG), and left coagulating gland (LCG).
The green two-headed arrow in the legend is there for the sake of completeness; there are no isomorphisms in this particular mapping. An isomorphism is a one-to-one and onto mapping between anatomical structures at a given granularity (in this case, Organ level) across species. In other words, each structure in one species at that level has one and exactly one correspondence in the other species. An example would be the heart, which is isomorphic not only at the Organ level: Heart (mouse) corresponds exactly to Heart (human), but also at the Chamber level: the human Left atrium, Left ventricle, Right atrium, and Right ventricle correspond exactly to those same structures in the mouse.
A null mapping (red bidirectional arrow) is the case when a structure exists only in one species and not at all in the other--for example, the human breast has no corresponding structure in fish models, because the breast is a structure found exclusively in mammals. So the Breast (human) maps to Null in fish.
A homomorphism (blue birdirectional arrow) is any relationship in between--it is a non-null mapping (so the structures exist in some form in both species), yet it is not one-to-one and onto (so there are structural differences of some kind which occurred during speciation). We see an example in the prostate here: the human prostate (a Lobular organ) corresponds in some way to 5 different lobular organs in the mouse, so the correspondence is 1:5 (or 5:1) rather than one-to-one and onto. It is, therefore, a homomorphism. Five blue bidirectional arrows are drawn to indicate the homomorphisms between the mouse and human organs.
There is a further homomorphism in this diagram; that between the Anatomical set comprised by the mouse organs--in other words, the mouse prostate in toto (MP)--and the human prostate in toto. Additionally, because there is no corresponding set of human prostates, there is a null mapping between the Anatomical set MP and the human (red bidirectional arrow).
The white unidirectional arrows indicate a subsumption relationship; the human prostate is-a Lobular organ, as is each organ of the mouse prostate. Further, the entire mouse prostate (MP) is-a Anatomical set. The membership (partitive) relationships (is-member and has-member) are conflated into a bidirectional dashed yellow arrow for the sake of clarity, as this diagram is already becoming quite complex, even at a level of granularity as gross as Organ.
Anatomical set is a species-independent anatomical abstraction, as is Lobular organ, indicated by the green circles. We do not map anatomical abstractions per se, as it does not make sense to say Anatomical set maps-to Lobular organ outside of any context, and as they are species-independent, there is no barrier to cross with a mapping.
What does make sense, however, is to map a structure in one species with a subsumption relationship to an anatomical abstraction to a structure in the second species with a subsumption relationship to a different anatomical structure. While there is no homomorphism directly between the anatomical abstractions themselves, the difference between the parents of the mapped structures is sufficient to indicate that an interesting anatomical transformation occurred during speciation. Therefore, although we do not map abstractions directly to other abstractions, we do keep track of subsumptions relationships of those structures which we do map.
These entities and relationships are tracked via the MAO, and similarities and differences among them are described using the SDM, and returned to the user via the CSAM interface.
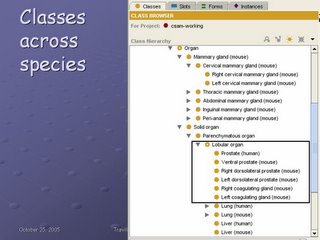
Because the anatomical abstractions are species-independent, there is only one anatomical taxonomy in Protege. For example, Lobular organ is defined once--it is-a Parenchymatous organ, which in turn is-a Solid organ--and it has as children not only the Prostate (human), but also all the mouse organs. Because there is only one taxonomy, the species is appended in parentheses at the end of the structure name.
CSAM extends the FMA, and as such it has access to the anatomical information in that ontology. It get slots such as adjacency, connectivity, blood supply, and innervation, among others, from the FMA.

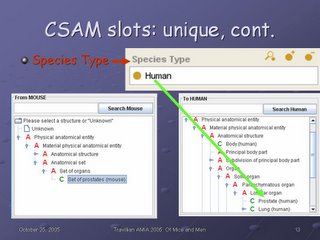
Additionally, it has slots which are unique to CSAM. One of the CSAM-specific slots, as mentioned previously, is the Relative name, where the species is appended in parentheses to the structure name. Another is the role, Abstract or Concrete, which distinguishes whether a structure can be mapped (Concrete), or--as an Anatomical abstraction--cannot, per our previous discussion
The green arrow shows how that information ends up in the CSAM interface: The red A in front of Lobular organ indicates an Anatomical abstraction, while the green C in front of Prostate (human) and Lung (human) indicates that those structures will support mapping queries.

Species Type is another CSAM-specific slot, and it is used to break the one species-independent hierarchy from Protege into two species-specific ones in the CSAM interface--the From hierarchy and the To hierarchy.
The last CSAM-specific slot is Maps-to. From this slot, the similarities--correspondences (isomorphism, homomorphism, null mapping)--and the differences (union, intersection, and set complement of result set) are calculated.
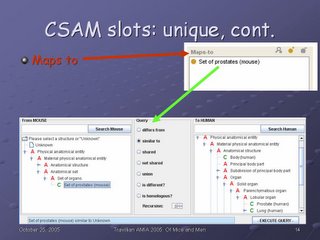
Queries currently supported include differs-from and similar-to. For example, "What structure in the mouse is similar to the lactiferous duct tree in the human?" returns the result set {Mammary gland (mouse)}. This result set is calculated via slot lookup and SDM, similarly to the way Emily functions.
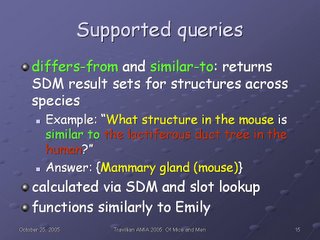
Is-different and is-homologous are the Boolean analogues of differs-from and similar-to: where the set queries ask for an unknown, the Boolean queries take two knowns and verify or dispute the proposed relationship.
Shared, not-shared, and union draw upon set and graph operations to answer more abstract queries: what structures are common across species, what structures are unique to one species or the other, and what are the range of possibilities for this structure in these species.

Our query syntax draws heavily from Emily: the query has three components, Subject, Relationship, and Object. Given any two in a query, CSAM can return the third component.
Like Emily, CSAM can use the result sets returned in previous queries as either the Subject or Object of further queries.
The range of allowable CSAM queries at this point comprise the ones we just reviewed.

That is the operation of the four sections of the CSAM interface: yellow box: specify directionality. Green box: store and retrieve previous query results. Blue box: display results of last submitted query (with tabs for optional tree display and display of associated graphics). Red box: specify a Subject-Relationship-Object query, enforcing correct syntax through the use of lists and radio buttons, and providing immediate feedback by translating the query into a text box.
At present, I am contacting domain experts (mouse anatomy and pathology experts for the sites we have identified) to establish what kinds of queries they would want in such a system to make it fit their information needs. We are adding content in accordance with their responses. Additionally, we are testing functionality as we add that content, and we project a future project is to extend this approach to answer queries about more MMHCC sites of interest.

I would like to thank all who made this work possible: the NLM, whose training grant funded this work; my committee; the domain experts, of whom this represents only a small sample, as they are too numerous to name individually; the programmers who have helped me to implement my design; and my mentor, Dr. Cornelius Rosse, the original developer of the FMA.


0 Comments:
Post a Comment
<< Home